48uep6bbphidcol2|ID
48uep6bbphidvals|1824
48uep6bbph|2000F98CTab_Articles|Fulltext
Gastrointestinal stromal tumors (GIST) commonly harbour oncogenic mutations of the proto-oncogene KIT tyrosine kinase. A subset of GIST, called ‘wild type’ possess mutations in Platelet derived growth factor receptor alpha (PDGFRA), most of which are resistant to the tyrosine kinase inhibior (TKI) imatinib. GIST having PDGFRA mutations are found most commonly in stomach. We report here a case of GIST presenting as an omental mass and being negative for CD117 (cKIT) on immunohistochemistry, which was further assessed by mutational analysis using next generation sequencing (NGS) and found to have PDGFRA mutation. This mutation, although silent, has been reported earlier in the literature; however it is the first case of its kind in Indian population.
Case Report
We report a case of 59 year-old, type II diabetic and hypertensive female, who presented with a history of dull aching abdominal pain, which was generalized, along with constipation. On per-abdomen examination, an 8 x 8 cm lump was palpable in left hypochondrium and left lumbar region, which was mobile, firm, non-tender and non-reducible. A contrast enhanced computed tomography imaging of abdomen revealed a large lobulated mass lesion, measuring 11.8 x 10.7 x 10.3 cm in left lumbar region, in close proximity to small bowel loops. The radiological opinion was highly suspicious for GIST/ mesenchymal tumor.
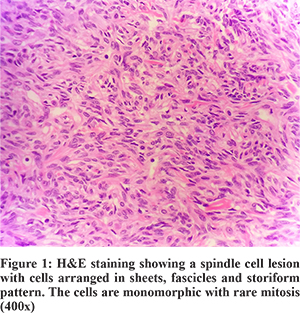
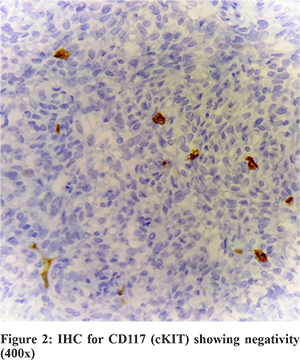
Patient underwent surgery for the same and the omental mass was sent for histopathological examination (Figure 1), which revealed a spindle cell lesion. This lesion was positive for CD34, bcl-2, CD99 and vimentin while lacking the expression for CD117 (cKIT), S100, SMA and EMA. In view of CD117 negativity (Figure 2), a mutational analysis was suggested for PDGFRA. The same was performed on the tissue obtained from histopathology. The DNA was obtained from the tissue block which was then used as the starting material for NGS.
Next generation sequencing was performed using the defined protocol, which included library preparation, template preparation and sequencing. The sequencing was performed on the Ion Torrent PGM platform. The analysis was performed using the Variant Caller plugin and also on the cloud-based Ion Reporter software. The sequencing showed the mutation in PDGFRA gene at locus chr4:55152040 (hg19), c.2472C>T, amino acid p.v824v, on exon 18 (Figure 3). The frequency of this mutation was 46% and depth of coverage was 1948x. This was a silent coding mutation. However, the sequencing of KIT gene did not reveal any mutation.
Discussion
The diagnosis of GIST has undergone a sea-of-change since their first recognition almost 30 years back. Its understanding has vastly improved with evolving diagnostic techniques especially molecular sequencing.
The common primary sites of GIST are the stomach (60%) and the small intestine (25–35%). Rarely GIST may be present in the esophagus or in the large bowel (<5%), the latter mostly in the rectum.1
GIST have a wide clinicopathologic spectrum in most sites ranging from minute incidental nodules to large tumors with variable gross appearances. Microscopic features are site dependent, but most GIST are spindle cell tumors, and a minority have epithelioid, mixed spindle, or rarely pleomorphic histology. A small minority of GIST(<5%) are negative for both CD34 and KIT, belonging to the subgroup driven alternatively by PDGFRA.2
GIST are characterized by expression of the type III receptor tyrosine kinase KIT in the vast majority of cases (>90%). More than 80% of GIST carry a gain of function mutation in the encoding region of KIT gene or alternatively in the PDGFRA gene leading to ligand independent autoactivation of one of the two tyrosine kinases.3
PDGFRA mutations are estimated to be present in 5-7% of GIST. PDGFRA gene mutation analysis help to predict responsiveness to the tyrosine kinase inhibitor (Imatinib) treatment.3 They are most often located in tyrosine kinase domain 2 encoded by exon 18. Point mutations are most common mutational subtype followed by deletions in the same region of the exon. Less often mutations are found in exon 12 and 14.
Mutations in PDGFRA gene can occur in various positions, of which the most common is at amino acid p.D842V (COSM12432), which is a missense mutation and is resistant to matinib. This is followed by another missense substitution p.V561D (COSM12395). Other less common mutations observed in GIST are p.I843_D846del, having deletion in frame (COSMIC28028), and p.N659K (COSMIC18893) having missense substitution. We report the case having silent substitution mutation of PDGFRA at amino acid p.V824V (COM22413).This kind of mutation is reported in very few cases and is probably first of its kind in Indian population. All the PDGFRA mutations except for p.D842V in exon 18 are sensitive to imatinib.
The mutational status of the tyrosine kinases, KIT and PDGFRA has become a well-established prognostic and predictive parameter for GIST. In metastatic or high-risk GIST it is essential to determine the mutational status as quickly as possible. Particularly in primary inoperable tumors mutational analysis is relevant for the clinical decision about a possible neoadjuvant therapy with imatinib.4 Therefore, multigene mutational analysis increasingly is performed on surgical specimens to help guide adjuvant therapy using Imatinib.4
Although there are similarities between PDGFRA mutant and KIT-mutant GISTs at the molecular level, a number of clinicopathologic differences in the specific cell morphology, metastasis, and prognosis have emerged in recent studies comparing these two ‘molecular variants’. In contrast to the KIT-mutant tumors, PDGFRA-mutant GIST more often have an epithelioid morphology, and more often have weak to negative of KIT (CD117) on immunohistochemistry. In addition, these tumors arise almost exclusively in the stomach, whereas KIT-mutant tumors occur at a variety of sites along the GI tract.5 Furthermore, the gene expression profiles of PDGFRA mutant tumors cluster separately from most KIT-mutant tumors. Thus, PDGFRA-mutant tumors represent a distinct subset of GIST that raise new challenges with regard to diagnosis (particularly those that are KIT negative) and treatment with TKIs.5
Our case was a rare presentation of GIST as omental mass. This lesion was immunohistochemically negative for routine markers like CD117, but had mutation of PDGFRA gene with V824V mutation which is sensitive for imatinib. DOG1 by immunohistochemistry is also a marker specific for GIST but was not available in our laboratory at that time. Although this is a silent coding mutation with no change in the amino acid, it is being reported for it being identified in the PDGFRA region in the CD117 negative GIST case. This mutation has been described in COSMIC (Catalogue of Somatic Mutation in Cancer) database with the id COSM22413 (c.2472C>T), and till date very few cases have been registered.
To conclude, performing the mutational analysis of GIST to identify mutations will not only be effective in diagnosis whenever there is histopathological dilemma of GIST but will also help in prognostication and treatment in view of imatinib sensitivity.
References
- Hirota S, Isozaki K. Pathology of gastrointestinal stromal tumors. Pathology International 2006; 56: 1–9.
- Wardelmann E, Büttner R, Merkelbach-Bruse et al. Mutation analysis of gastrointestinal stromal tumors: increasing significance for risk assessment and effective targeted therapy: Virchows Arch 2007; 451:743–749.
- Gleeson FC, Kipp BR, Kerr SE et al. Kinase Genotype Analysis of Gastric Gastrointestinal Stromal Tumor Cytology Samples Using Targeted Next-Generation Sequencing. Clinical Gastroenterology and Hepatology 2015; 13(1): 206-6.
- Ku¨nstlinger H, Huss S, Merkelbach-Bruse S et al. Gastrointestinal Stromal Tumors With KIT Exon 9 Mutations Update on Genotype-Phenotype Correlation and Validation of a High- Resolution Melting Assay for Mutational Testing; Am J Surg Pathol 2013;37:1648–1659
- Miettinen M, Lasota J. Gastrointestinal stromal tumors–definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1–12.