|
Stella I. Smith, Emmanuel A. Omonigbehin, Helen A. Goodluck, Fatimah B. Abdulkareem2, Charles A. Onyekwere3, Chimere Agomo4, Dennis A. Ndububa5, Muinah A. Fowora, Jesse A. Otegbayo6, Monica Contreras7, Rainer Haas8, Gabriele Rieder8
*Molecular Biology and Biotechnology Division,
Nigerian Institute of Medical Research (NIMR),
Nigeria, Dept of Morbid Anatomy,
College of Medicine,
University of Lagos,
Nigeria,2
*Dept. of Medicine, Lagos State University Teaching Hosptital,
Nigeria,3
* Biochemistry and Nutrition Division,
Nigerian Institute of Medical Research,
Nigeria,4
* Dept. of Medicine, Obafemi Awolowo University Teaching Hospital Complex,
Ile-Ife, Ife, Nigeria,5
* Dept. of Medicine, University College Hospital, Ibadan,
Nigeria6
* Venezuelan Institute for Scientific Research (IVIC),
Caracas, Venezuela,7
* Max von Pettenkofer Institute,
LMU, University of Munich,
Germany8
Corresponding Author:
Dr. SI Smith
Email: stellaismith@yahoo.com
DOI:
http://dx.doi.org/20862986
48uep6bbphidvals|325 48uep6bbph|2000F98CTab_Articles|Fulltext Helicobacter pylori is a curved, highly motile, gram-negative rod that colonizes the mucosa of the human stomach of more than half of the world’s population.[1,2] Infection is mainly acquired in childhood and stays asymptomatic for decades in approximately 80% of cases.[2] There are several diagnostic tests for the detection of H. pylori and are grouped into two categories: (I) invasive tests that require endoscopy such as culture, rapid urease test (RUT), histology, direct gram-stain of biopsies, and PCR; (II) non-invasive or minimally invasive tests such as serology, urea breath test (UBT), and H. pylori stool antigen tests (HpSA).[3]
Although culture is the gold standard, it is very much dependent on some infrastructure conditions and power supply. Therefore, the culturing of H. pylori can be unreliable especially for an environment like Nigeria where power outages are quite regular and lasting for days. This is also in addition to the problem of overgrowth or contamination with other microorganisms. Due to this situation, cultivation of H. pylori is not routinely applied for the detection of H. pylori in Nigeria. However, culture is necessary in treating cases with failure of primary attempt at H.pylori eradication. The study aimed at evaluating various methods for the diagnosis of H. pylori in developing countries where facilities for culture of H.pylori may not be easily available at all centres.
Methods
167 patients with dyspepsia undergoing upper gastrointestinal endoscopy were included after informed consent. Patients on NSAIDs, PPIs, and antibiotics were excluded. A total of 835 stomach biopsy samples (three antrum and two corpus) were obtained from these patients during endoscopy. The study was carried out between January 2008 and December 2008. The biopsies were subjected to H. pylori culture, CLO test kit (Delta West pty, Australia), H. pylori stool antigen test (HpSA, Meridien UK) using stool samples or rectal swabs, serology test (Human, Germany), histology (Giemsa stain), and direct gram stain of smear from biopsy. Three biopsies from the antrum were used :- one each for CLO test, culture/ PCR, and histology. While the other two corpus biopsies were used only for histology and culture/PCR. All tests with kits were carried out according to their manufacturer’s instructions. The specimens were obtained from two centres in Nigeria: Lagos State University Teaching Hospital (LASUTH, Ikeja) and Obafemi Awolowo University Teaching Hospital Complex (OAUTHC), Ile Ife. Ethical approval was obtained from the NIMR-IRB before the study commenced.
Direct detection of H. pylori DNA from gastric biopsy samples
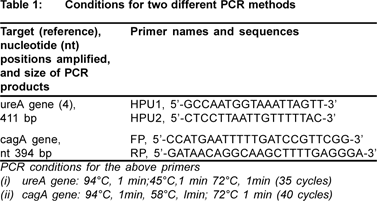
DNA from biopsies was extracted by using the method of Marais et al.4 Primers and cycling parameters used in this study for amplifying the genes ureA (411bp) and cagA (394bp) are documented in Table 1.
Specificity of PCR assay
The specificity of PCR primers targeting the ureA and cagA genes were determined by testing other bacterial strains (Salmonella sp.) from related genus.
Statistical analysis
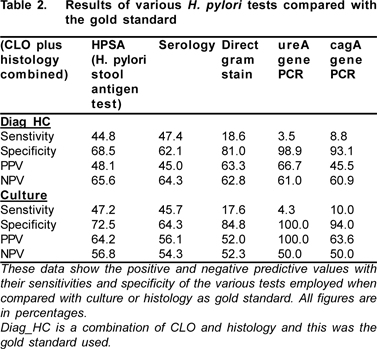
The results were analyzed by chi-square test. Results are presented as mean ± standard deviation for quantitative variables and number (percentages) for qualitative variables. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were measured using the culture as well as histology as gold standard.
Results
A total of 167 patients participated in this study: 46 (27.5%) from Ife, Osun State and 121 (72.5%) from Ikeja, Lagos state. The participants of this study had a mean age of 46.6±15.9 years (range 10-100 years). Most of them (47.7%) fell into the age group of 30-50 years. The sex distribution was even with 83 (49.7%) being females and 84 (50.3%) males. There was no significant difference in the age and sex distribution of the study participants from the two sites used for this study.
Endoscopic findings
Upper gastrointestinal endoscopy findings were normal, gastric ulcer, duodenal ulcer, and reflux oesophagitis in 19.9%, 15.7%, 13.2%, and 7.8% of patients respectively. H.pylori positivity as seen by various diagnostic tests was: 37.1% - CLO test, 36.7%-HpSA, 48.7% -culture, 41.7% - serology, 18.9%-direct Gram stain , 32.7% - histology , 2.1% - ureA PCR, 7.6% - cagA PCR.
Specificity of PCR assays with bacterial isolates
The specificty of the PCR primers targeting ureA and cagA genes was determined by testing non-H. pylori-strains. The Salmonella sp. tested were negative for the various genes tested.
Using sensitivity as a measure of the ability of the various methods to detect the presence of H. pylori infection as compared with culture and histology, HpSA and blood serology were the best methods for detecting H. pylori (Table 2).
Discussion
From our study, the prevalence of H. pylori by culture was found to be 48.7%. This is an increase in the prevalence reported previously by Smith et al[5] of 27%. In other reports from developing countries, e.g. in a pilot study from the Philippines, H. pylori culture was 26%.[6] It has also been reported that the prevalence of infection is greater in developing countries and is influenced by socioeconomic conditions, ethnic background, and age.[7,8] The power outages in Nigeria have made the culturing of H. pylori unreliable. Together with the risks of overgrowth or contamination it appears to be the least sensitive and most cumbersome method for H. pylori detection in Nigeria .. Another gold standard commonly used for the diagnosis of H. pylori is histology. From our study the prevalence of H. pylori using histology was 32.7% and in a few cases where histology was negative one or two other tests were positive for H. pylori. This was corroborated by some reports where it was reported that a false negative result by histology of up to 25% was also possible, especially if a special stain (Diff-Quik) was not included.[3] Another report by Destura et al[6] showed a similar comparison with histology, culture, and CLO test H. pylori can be readily detected at endoscopy by histology, culture or urease tests.
Using the CLO test, 37.1% of the patients revealed H. pylori infection in this study. Although CLO test is highly specific for H. pylori infection, it requires high density of bacteria, nevertheless it is commonly used for the detection of H. pylori infection at endoscopy.[3] The sensitivity of urease test is reduced in patients who are taking proton pump inhibitors (PPI), antibiotics or bismuth compounds.[3] Therefore, we excluded those patients. Any antibiotic active against H. pylori will cause a reduction in the numbers of bacteria in the stomach.[3] In Nigeria, across over the counter purchase of PPIs is a common phenomenon. It could be possible that some of the patients had taken self-medication and did not state it during the initial survey.
The prevalence of H. pylori from our study using the HpSA was 36.7% and that of serology was 41.7%. These two diagnostic tests produced the highest prevalence rate in our study. Reports have indicated the accuracy of HpSA in detecting H. pylori infections even as a screening test for post treatment and epidemiological purposes.[3] It is especially useful when a large scale epidemiological study for H. pylori acquisition is carried out in children.[3] Although serology is good for epidemiological purposes, it might not be useful for post eradication therapy as the antibodies remain long after eradication of infection.[3]
The direct Gram stain was one of the least sensitive methods with a prevalence of 18.9%. It could be attributable to the fact that maybe as result of low numbers of H. pylori, the spiral shape was not identified by microscopy. In addition, the assessment is also subjective and could be due to human error.
From this study the positive predictive value (PPV) of the two molecular tests (ureA and cagA gene) with culture as the gold standard was 100% and 63.6%, respectively. This is similar to a previous report by Smith et al[9] which showed the PPV of ureA to be 91% using culture as the gold standard. In comparison, negative predictive values (NPV) from this study was 50% each while previous study showed at least 44%. In terms of specificity, for both methods being used as gold standards (culture and histology) the molecular based methods were more specific than the other methods although, the sensitivity was however the opposite. False negative results from this study could have occurred due to low numbers of H. pylori. In a recent report by Sugimoto et al,[10] it was concluded that PCR currently used for the direct detection of H. pylori in clinical or environmental samples were unreliable. This is due to many false positives obtained and suggested the possible use of identification based on the presence of multiple putative virulence factor genes.
In conclusion, data from Nigeria suggests that a combination of stool antigen tests and serology had the highest diagnostic accuracy in detecting H.pylori infection.
Acknowlegments
The authors acknowledge the support of CRP-ICGEB (International Centre for Genetic Engineering and Biotechnology,grant no CRP/NIG07-02) and NIMR (Institute Nigerian Institute of Medical Research) for sanctioning grant for this research work.
References
1. Blaser MJ. Ecology of Helicobacter pylori in the human stomach. J Clin Invest. 1997;100:759–62.
2. Neale KR, Logan RP. The epidemiology and transmission of Helicobacter pylori infection in children. Aliment Pharmacol Ther. 1995;9:77–84.
3. Graham KS and Graham DY. Contemporary diagnosis and Management of H. pylori associated diseases. 2nd Edition. Published by Handbooks in Health Care Co, Newtown, Pennsylvania, USA. 2002.p.70–95.
4. Marais A, Monteiro L, Occhialini M, Pina M, Lamouliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–7.
5. Smith SI, Oyedeji KS, Arigbabu A, Anomneze EE, Chibututu CC, Atimomo CA, et al. Prevalence of H. pylori in patients with gastritis and peptic ulcer in Western, Nigeria. Biomedical Lett. 1999;60:115–20.
6. Destura RV, Labio ED, Barrett LJ, Alcantara CS, Gloria VI, Daez ML, et al. Laboratory diagnosis and susceptibility profile of Helicobacter pylori infection in the Philippines. Ann Clin Microbiol Antimicrob. 2004;3:25.
7. Moayyedi P, Axon AT, Feltbower R, Duffett S, Crocombe W, Braunholtz D, et al. Relation of adult lifestyle and socioeconomic factors to the prevalence of Helicobacter pylori infection. Int J Epidemiol. 2002;31:624–31.
8. Adeyemi EO, Danial MF, Helal T, Benedict S, Abdulle AM. The outcome of a 2-week treatment of Helicobacter pylori-positive duodenal ulcer with omeprazole-based antibiotic regimen in a region with high metronidazole resistance rate. Eur J Gastroenterol Hepatol. 1999;11:1259–63.
9. Smith SI, Oyedeji KS, Arigbabu AO, Cantet F, Megraud F, Ojo OO, et al. Comparison of three PCR methods for the detection of Helicobacter pylori DNA and detection of cagA gene in gastric biopsy specimens. World J Gastroenterol. 2004;10:1958–60.
10. Sugimoto M, Wu JY, Abudayyeh S, Hoffman J, Brahem H, Al- Khatib K, et al. Unreliability of Results of PCR Detection of Helicobacter pylori in clinical or environmental samples. J Clin Microbiol. 2009;47:738–42.
|