|
|
|
|
 |
 |
| |
 |
|
|
Original Articles |
|
|
|
|
|
Keywords :
Hepatitis C virus, IL28B, genetic association, genotype 3 HCV |
|
|
Sreedhar Chinnaswamy,1 Kausik Das,2 Bijan B Bairagya,1 Chandrika Bhattacharyya,1 Shalimar,3 Ajay Duseja,4 Deepak Amarapurkar,5 Rosangluaia,6 Sujit Chowdhury,7 Ashokanand Konar,8 Partha P Majumder,1 Abhijit Chowdhury2
National Institute of Biomedical
Genomics, PO - NSS,
2nd Floor, Kalyani, West Bengal,1;
Division of Hepatology, School of
Digestive and Liver Diseases, Institute
of Post Graduate Medical Education
and Research, Kolkata,2;
Division of Gastroenterology and
Human Nutrition, All India Institute
of Medical Sciences, New Delhi3;
Department of Hepatology,
Postgraduate Institute of Medical
Education and Research, Chandigarh4;
Department of Gastroenterology,
Bombay Hospital and Medical
Research Centre, Mumbai5;
Civil HospitalAizawl, Mizoram6;
AMRI Hospital, Salt Lake, Kolkata7;
Peerless Hospital, Kolkata8;
India
Corresponding Author:
Dr. Sreedhar Chinnaswamy
Email: sc2@nibmg.ac.in
Dr. Abhijit Chowdhury
Email: achowdhury2002@yahoo.
co.in
DOI:
http://dx.doi.org/10.7869/tg.187
Abstract
Background and aim: IL28B gene polymorphisms have been associated with treatment-response (sustained virological response, SVR) in genotype 1 hepatitis C virus (HCV)-infected patients, but only with early phase of viral decline (rapid virological response, RVR) with genotype 3 HCV-infected patients. Association between IL28B variants and SVR in genotype 3 HCVinfected patients is unclear. Our study aimed to replicate the association of IL28Bsingle nucleotide polymorphism (SNP) rs8099917 with SVR and to validate its association with RVR in genotype 3 HCV-infected patients.
Methods: 72 patients receiving combination therapy (interferon-alpha and ribavirin) at different Indian centers were retrospectively recruited and their genotype at rs8099917 was determined. The association with RVR and SVR was tested taking in to account the variation in relevant covariates such as age, gender, baseline HCV RNA copy number and liver enzymes.
Results: The minor allele frequency (MAF) in the pooled samples was 0.17 at rs8099917 (G allele). 68% had TT, 29% had GT and 3% had the GG genotype. SVR was achieved in 71% of patients. A significant association of rs8099917 with both RVR (p=0.026) and SVR (p=0.016) was observed with none of the covariates showing any significant association. The relapse rate was high (20%) but no association of rs8099917 was observed with relapse (p=0.420).
Conclusion: An IL28B SNP associates with both early phase of viral decline and sustained response in a cohort of genotype 3 HCV-infected patients from India.
|
48uep6bbphidvals|646 48uep6bbph|2000F98CTab_Articles|Fulltext Hepatitis C virus (HCV) is a global public health problem[1] and the burden of chronic liver disease due to HCV is significant not only in Europe and America but also in south Asia, including India.[2-5] Pegylatedinterferon and ribavirin (PR) therapy forms the standard of care worldwide, but is expensive and has side-effects that often necessitates dosage adjustments.The response rate to PR therapy defined as sustained test-negativity for serum HCV RNA for at least six months after termination of therapy measured as sustained virological response (SVR). Varies between 40-70% depending on the viral genotype.[6,7] Further, the viral genotype shows significant geographical variation.[8] The difficult-to-treat genotype 1 predominates in the West, but genotypes 2 and 3 which respond much better to standard therapy are more prevalent in Asia, including India.[2,5] Several pre-treatment and on-treatment predictors of response to PR therapy including demography, disease status, viral load and genotype, have been used to individualize treatment and duration of therapy.[9-13] Recently, however, there has been a paradigm shift in pretherapy predictors of response to therapy in chronic HCV infections. Single nucleotide polymorphisms of the IL28B gene on chromosome 19 encoding interferon lambda have emerged as an important determinant of treatment response and spontaneous clearance of HCV.[14-18] All the studies that led to this discovery involved difficult-to-treat genotype 1 HCVinfected patients where individualization of treatment had been a significant unmet need. Subsequent studies have successfully validated these findings in both genotype 1[19]and genotype[2] and 3 infected patients from different ethnic groups.[20-22] There are differences in the strength of the association as well as predicted relationship to SVR depending on the viral genotype. Association studies on genotypes 2 and 3 have shown inconsistent results so far.[15-18,20,23-25] Relapse after therapy is an important concern in genotype 3 HCVinfection.[20,25] It has been suggested the duration of therapy may need to be individualized in genotype 3 infections.[26,27]Importantly, the association of IL28Bpolymorphismswith treatment outcome in genotype 3 HCV patients from India has not been studied. Therefore, we examined the association of rs8099917 in a cohort of Indians infected with genotype 3 HCV who received standard combination therapy. We chose this SNP becauseit has been strongly associated with treatment response in an Asian genome-wide association study[17] The rs8099917 polymorphis has been found to be a better predictor of treatment response in Asiansthan other IL28B SNPs.[28]A recent study from Japan has shown that treatment response (both RVR: rapid virological response, defined as conversion to HCV-RNA test-negativity after 4 weeks of therapy and SVR) in a cohort of genotype 2
HCV-infected patients was significantly associated with rs8099917.[29] Other studies[30,31] have also demonstratedthe association of IL28B SNPs with SVR in genotype 2/3 HCV infections. However, none of these studies have exclusively examined this SNP in genotype 3 HCV patients.Besides these three studies, others have failed to show any significant association betweenIL28B polymorphisms and SVR in genotypes 2 and 3 HCV-infected patients[32] although all studies have shown an association with RVR. A recent report also shows significant association between a novelIL28B SNP (ss469415590; encoding IFN-L4) with SVR[33] in genotype 2/3 HCV infections.Therefore, the association of IL28B SNPs and SVR in genotype 2/3 HCV infections remains controversial. The present study shows that the rs8099917 SNP is associated with both RVR and SVR in genotype 3 patients, though the association is weak and shows no association with relapse.The latter is a relatively novel finding, and raises the need for larger studies to validate the role of IL28B polymorphism singenotype 3 treatment outcome.
Methods
Study design
This was a retrospective, multicentric study involving previously PR treated genotype 3 chronic HCV-infected patients from seven (three from eastern India, one from north-east, two from north and one from west) centers across India. Patients who received PR therapy between 2009 and 2012 were recruited and their blood samples were drawn for SNP genotyping. The protocol including consent forms and sharing of patient medical records was approved by the Institutional Ethics Committee - Review Committee for Protection of Research Risks to Humans, National Institute of Biomedical Genomics, Kalyani.
Patients
Seventy-two genotype 3 chronic HCV patients (median age: 40 years; 69% males) were enrolled in the study. All of them had previously completed a course of pegylated interferon and ribavirin therapy. Data on demography, clinical status, liver function tests, hematological indices, duration and dosage of treatment, interruptions and discontinuation of therapy were all recorded and pooled from all participating centers. Viral load at different time points including RVR, EVR, SVR, end of treatment response and at follow up (current and last available in case of relapse) were also recorded. EVR, denotes the early virological response defined as a minimum log-2 decline in HCV RNA after 12 weeks of therapy. Viral load was estimated at each Institute by standardized real-time PCR protocols. Patients were excluded if they had co-infection with HIV or HBV, had other co-morbidities, and suffered from alternate causes of liver disease like alcoholism.
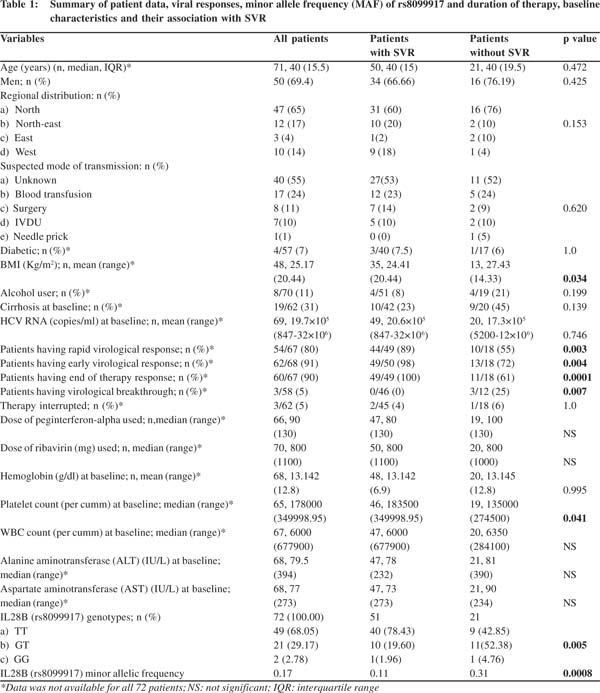 Genotyping of rs8099917 from genomic DNA
Genomic DNA was isolated from whole blood using the QiagenMidiPrep kit as per the manufacturer’sinstructions. The isolated DNA was electrophoresed on 1% agarose gels for quality check and the DNA was quantified using Nanodrop (Thermo Scientific). The SNP rs8099917 was genotyped by RFLP. PCR was carried out using the primers: a) 917FOR700bp (5’CAAGTCAGGCAACCACATGC3’),and b) 917REV 700bp (5’GCTCTGTCTGTCTCAATCAATC-3’). Each reaction included 200-400ng of genomic DNA as template in a total reaction volume of 20 µl. Each primer was added to a final concentration of 0.8 µmol. Taq polymerase (New England BioLabs, 5x master mix) was used for DNA amplification. The PCR conditions were as follows: initial denaturation for 5 minutes at 94oC; denaturation for 1 minute at 94oC, annealing for 45 seconds at 58oC, extension for 45 seconds at 72oC and these steps were repeated for 30 cycles with a final extension of 10 minutes at 72oC. Determination of rs8099917 genotype was carried outby RFLP of the 730 bp amplicongenerated in the previous step. The amplified DNA was incubated overnight(12-18 hours at 65oC) with the restriction enzyme BsrDI (New England BioLabs).The restriction-digested DNA was run on 2% agarose gel.A 100 base pair ladder (Fermentas) was run in parallel to determine the sizes of the digested fragments.
Statistical analysis
Fisher’s exact test was used for testingthe associationbetween SVR and rs8099917. Chi-square test, Mann-Whitney U- test, and student’s T-test were used for other comparisons of SVR with different clinical parameters.A binomial logistic regression model was used to test the association of RVR and SVR with rs8099917 with age, gender, alanine aminotransferase (ALT) andaspartate aminotransferase (AST) levels as covariates. A dominant model which tested patients with at least one copy of the minor allele as one group, against those with two copies of the major allele was used. A p value <0.05 was taken as significant.
Genotyping of rs8099917 from genomic DNA
Genomic DNA was isolated from whole blood using the QiagenMidiPrep kit as per the manufacturer’sinstructions. The isolated DNA was electrophoresed on 1% agarose gels for quality check and the DNA was quantified using Nanodrop (Thermo Scientific). The SNP rs8099917 was genotyped by RFLP. PCR was carried out using the primers: a) 917FOR700bp (5’CAAGTCAGGCAACCACATGC3’),and b) 917REV 700bp (5’GCTCTGTCTGTCTCAATCAATC-3’). Each reaction included 200-400ng of genomic DNA as template in a total reaction volume of 20 µl. Each primer was added to a final concentration of 0.8 µmol. Taq polymerase (New England BioLabs, 5x master mix) was used for DNA amplification. The PCR conditions were as follows: initial denaturation for 5 minutes at 94oC; denaturation for 1 minute at 94oC, annealing for 45 seconds at 58oC, extension for 45 seconds at 72oC and these steps were repeated for 30 cycles with a final extension of 10 minutes at 72oC. Determination of rs8099917 genotype was carried outby RFLP of the 730 bp amplicongenerated in the previous step. The amplified DNA was incubated overnight(12-18 hours at 65oC) with the restriction enzyme BsrDI (New England BioLabs).The restriction-digested DNA was run on 2% agarose gel.A 100 base pair ladder (Fermentas) was run in parallel to determine the sizes of the digested fragments.
Statistical analysis
Fisher’s exact test was used for testingthe associationbetween SVR and rs8099917. Chi-square test, Mann-Whitney U- test, and student’s T-test were used for other comparisons of SVR with different clinical parameters.A binomial logistic regression model was used to test the association of RVR and SVR with rs8099917 with age, gender, alanine aminotransferase (ALT) andaspartate aminotransferase (AST) levels as covariates. A dominant model which tested patients with at least one copy of the minor allele as one group, against those with two copies of the major allele was used. A p value <0.05 was taken as significant.
Results
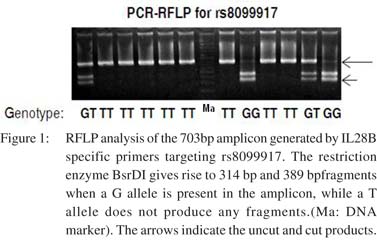
Genotypes at rs8099917 are significantly associated with SVR in genotype 3 HCV-infected patients The study population included only genotype 3 (subtype a or b) HCV-infected patients enrolled from different Indian centers. The 72 patients hailed from different geographical regions with majority being north Indians (Table 1). Their median age was 40 years and 69% were males. SVR was achieved in 71% patients. The rs8099917 SNP is associated with treatment response in Asians with genotype 2HCV infections.28,29RFLP analysis yielded unambiguous genotypes for the rs8099917 SNP (Figure 1). The minor allele frequency (MAF) was 0.17 at rs8099917 (G allele). 68% had TT genotype, 29% had GT and 3% had the GG genotypes (Table 1). Chi-square test on the genotyping data confirmed that the population was in Hardy- Weinberg equilibrium (p>0.05). We tested the TT genotype versus non-TT genotype for association with SVR using Fisher’s exact test. We found that rs8099917 showed significant association with SVR (p=0.005), with 42.8% patients with TT genotype and 57.1% patients with a non-TT genotype in the non-SVR group (Figure 2). We performed statistical tests to detect association of physical, biochemical and clinical parameters with attainment of SVR (Table 1). In brief, apart from RVR, EVR, end of treatment response and virological breakthrough we found a weak association with baseline platelet counts (p=0.041) and BMI (p=0.034).
 Predictors of response
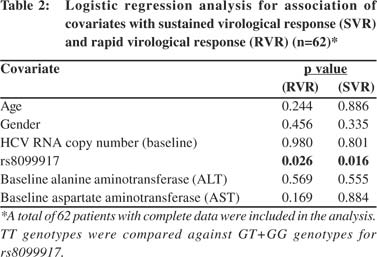
Further analysis using a binary logistic regression model taking age, gender, AST, ALT and baseline HCV RNA copy number as covariates showed significant association of rs8099917 with SVR (odds ratio: 4.42; 95% CI: 1.36 to 14.39, p=0.016) (Table 2).
Predictors of response
Further analysis using a binary logistic regression model taking age, gender, AST, ALT and baseline HCV RNA copy number as covariates showed significant association of rs8099917 with SVR (odds ratio: 4.42; 95% CI: 1.36 to 14.39, p=0.016) (Table 2).
None of the covariates showed any significant association. Since a weak association between baseline platelet count and treatment response was noted (Table 1), its inclusion as a covariate did not alter the significant association between SVR and rs8099917. We next tested the association of rs8099917 with RVR. 81% of the patients showed RVR and the TT genotype showed significant association with RVR (odds ratio: 5.69; 95% CI:1.47 to 22.10,p=0.026) compared to non-TT genotype with none of the covariates influencing the observed association (Table 2).
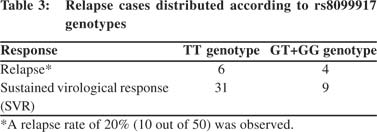
Genotypes at rs8099917 do not significantly associate with relapse Earlier studies on genotype 3 HCV-infected patients have shown association with RVR but not SVR, due to high relapse rates.20,25 In our cohort even though the relapse rate was as high as 20% (Table 3) there was no association of rs8099917 with relapse (p=0.420). These results suggest that similar to genotype 1 HCV-infectedpatients, the genotype 3 HCV-infected patients from our population show association at an IL28B SNP with both RVR and SVR.
Discussion
The association of IL28B SNPs with treatment response in genotype 2 and 3 HCV-infected patients has been controversial. Several studies have shown association of IL28B SNPs with RVR in genotype 3 HCV infections, but no significant association has been seen with SVR.20,24,25 Mangia et al from Italy genotyped ~300 chronic HCV genotype 2/3 infected patients who received standard combination therapy. They were tested for associationsbetween rs12979860 genotypes and RVR and SVR. The C allele at the SNP was associated with SVR in only those patients who did not show RVR. In the patients who showed RVR, SVR did not depend on the IL28B genotype. This is in contrast to genotype 1 HCV patients in whom SVR is significantly associated with rs12979860 irrespective of the RVR status.[34] Scherzer et al from Austria did not find any significant association of IL28B polymorphisms rs12979860 and rs8099917 with SVR in genotype 3 HCV-infected patients.[24] However, early response to treatment was significantly associated with the two SNPs, with the CC genotype and TT genotype, respectively, favoring a faster viral decline during weeks 2-4 of therapy.[24] In another study from Norway and Denmark involving genotype 3 HCV-infected patients, RVR was achieved by a significantly greaternumber of patients who had CC and TT genotypes at rs12979860 and rs8099917, respectively, but these genotypes showed no association with SVR.[20] The study also found that CC genotype at rs12979860 and TT genotype at rs8099917 were associated with greater liver damage (measured by markers like ALT) and higher baseline viral load.[20] The above studies suggest that genotype 2 and 3 HCV infections may have different mechanisms of immune activation. The IL28B SNPs appear to be associated with early viral decline but not with the subsequent sustained response, although no consensus is available on the issue. Interestingly Sakamoto et al have shown a significant association of rs8099917 with both RVR and SVR in genotype 2 HCV-infected patients, although the association was observed with only genotype 2b HCV infections.[29]
Sarrazin et al reported that an IL28B SNP is associated with SVR only in genotype 2/3 HCV patients who have already achieved an RVR23 However, Moghaddam et al have shown a high relapse rate within the RVR group comprising of genotype 3 patients carrying the favorable IL28B genotypes.[20] A recent report from the United Kingdom involving genotype 3 HCV infected patients also reported similar findings.[25] Several explanations can be offered for the above observations. Firstly, these studies suggest that genotype 3 HCV infections may have reservoirs that are resistant to the combination therapy and lead to relapse. Secondly, due to high response rates in genotype 3 HCV-infected patients, studies with this HCV genotype may be grossly underpowered to detect a small effect size from the Il28B gene.[35] A third possible explanation pertains to the duration of combination therapy in genotype 3 HCV infections. A 24-week therapy is usually considered sufficient for achieving an SVR in genotype 3 HCV-infected patients.But no study has examined the effect of longer therapy in achieving SVR in light of the IL28B SNPs. Such studies can elucidate how genotype 3 HCV infections are clinically and pathologically different from genotype 1 infections.
HCV infection is becoming a major public health problem in India with a prevalence as high as 5% in some parts of the country.[4] Genotype 3 HCV predominates in India and Pakistan as opposed to genotype 1 HCV in the Western world.[5] The reason for this difference is unclear. We examined the association of IL28BSNPs with treatment response in genotype 3 HCV-infected Indian patients. We detected a statistically significant association between rs8099917 and SVR in our cohort of 72 patients. The association persisted even after adjusting for relevant covariates like HCV RNA copy number, age, gender and liver enzyme levels. We also observed similar statistically significant association with RVR.
Our results suggest that even though genotype 3 HCV infections respond better to combination therapy, the response may involve innate immune mechanisms similar to those against genotype 1 HCV infection.[36] Our results for RVR are similarto other studies on genotype 3 HCV infections, but our results with SVR are novel. We also found that relapse has no association with IL28B genotype (Table 3), unlike previous studies where IL28B genotype-associated relapse explained the lack of association with SVR.[20,25] Three previous studies have showed significant association between SVR and IL28B SNPs,[29-31] although these studies involved only genotype 2 or genotype 2/3 HCV patients. Furthermore, a recent meta-analysis has demonstrated that genotype 2/3 HCV association studies carried out so are limited by their sample size.[35] The meta-analysis shows that favorable IL28B genotypes are significantly associated with both RVR and SVR in genotype 2/3 HCV-infected patients, although the impact of IL28B polymorphisms on a sustained response is limited in these cases.[35] Our resultswhen combined with previous reports[29-31] suggest that genotype 2 and 3 HCV replication in humans is affected by IL28B polymorphisms similar to the effects observed on genotype 1 HCV replication.[36,37]
In summary, we report that the TT genotype at the rs8099917 SNP is significantly associated with both rapid and sustainedvirological response in Indian genotype 3 HCVinfected patients.The lack of IL28B genotype-dependent relapse may be responsible for association of rs8099917 with both RVR and SVR in our cohort. Larger studies areneeded to validate our findings in genotype 3 HCV Indian patients.
References
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558-67.
- Mukhopadhyaya A. Hepatitis C in India. J Biosci. 2008;33:465–73.
- Chowdhury A, Santra A, Chaudhuri S, Dhali GK, Chaudhuri S, Maity SG, et al. Hepatitis C virus infection in the general population: a community-based study in West Bengal, India. Hepatology. 2003;37:802–9.
- Sood A, Sarin SK, Midha V, Hissar S, Sood N, Bansal P, et al. Prevalence of hepatitis C virus in a selected geographical area of northern India: a population based survey. Indian J Gastroenterol. 2012;31:232–6.
- Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, et al. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31 Suppl 2:61–80.
- Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr., et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82.
- Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, et al. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993–9.
- Sy T, Jamal MM. Epidemiology of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:41–6.
- Davis GL, Lau JY. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26:122S–7S.
- McHutchison JG, Shad JA, Gordon SC, Morgan TR, Ling MH, Garaud JJ, et al. Predicting response to initial therapy with interferon plus ribavirin in chronic hepatitis C using serum HCV RNA results during therapy. J Viral Hepat. 2001;8:414–20.
- Fried MW. Viral factors affecting the outcome of therapy for chronic hepatitis C. Rev Gastroenterol Disord. 2004;4:S8–S13.
- Berg T, Sarrazin C, Herrmann E, Hinrichsen H, Gerlach T, Zachoval R, et al. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37:600–9.
- Manns M, Zeuzem S, Sood A, Lurie Y, Cornberg M, Klinker H, et al. Reduced dose and duration of peginterferon alfa-2b and weight-based ribavirin in patients with genotype 2 and 3 chronic hepatitis C. J Hepatol. 2011;55:554–63.
- Doyle JS, Hellard ME, Thompson AJ. The role of viral and host genetics in natural history and treatment of chronic HCV infection. Best Pract Res Clin Gastroenterol. 2012;26:413–27.
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatmentinduced viral clearance. Nature. 2009;461:399–401.
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–4.
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9.
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801.
- Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV—response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9:406–17.
- Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjoro K, et al. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746–54.
- Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, et al. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821–7, 7 e1.
- Lindh M, Lagging M, Farkkila M, Langeland N, Morch K, Nilsson S, et al. Interleukin 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 or 3 of hepatitis C virus. J Infect Dis. 2011;203:1748–52.
- Sarrazin C, Susser S, Doehring A, Lange CM, Muller T, Schlecker C, et al. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415–21.
- Scherzer TM, Hofer H, Staettermayer AF, Rutter K, Beinhardt S, Steindl-Munda P, et al. Early virologic response and IL28B polymorphisms in patients with chronic hepatitis C genotype 3 treated with peginterferon alfa-2a and ribavirin. J Hepatol. 2011;54:866–71.
- Bucci C, von Delft A, Christian A, Flemming VM, Harrison A, Halliday J, et al. ‘Favourable’ IL28B polymorphisms are associated with a marked increase in baseline viral load in hepatitis C virus subtype 3a infection and do not predict a sustained virological response after 24 weeks of therapy. J Gen Virol. 2013;94:1259–65.
- Liang TJ. Shortened therapy for hepatitis C virus genotype 2 or 3—is less more? N Engl J Med. 2007;357:176–8.
- Dalgard O, Bjoro K, Hellum KB, Myrvang B, Ritland S, Skaug K, et al. Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot study. Hepatology. 2004;40:1260–5.
- Ito K, Higami K, Masaki N, Sugiyama M, Mukaide M, Saito H, et al. The rs8099917 polymorphism, when determined by a suitable genotyping method, is a better predictor for response to pegylated alpha interferon/ribavirin therapy in Japanese patients than other single nucleotide polymorphisms associated with interleukin-28B. J Clin Microbiol. 2011;49:1853–60.
- Sakamoto N, Nakagawa M, Tanaka Y, Sekine-Osajima Y, Ueyama M, Kurosaki M, et al. Association of IL28B variants with response to pegylated-interferon alpha plus ribavirin combination therapy reveals intersubgenotypic differences between genotypes 2a and 2b. J Med Virol. 2011;83:871–8.
- McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307–14.
- Cavalcante LN, Abe-Sandes K, Angelo AL, Machado TM, Lemaire DC, Mendes CM, et al. IL28B polymorphisms are markers of therapy response and are influenced by genetic ancestry in chronic hepatitis C patients from an admixed population. Liver Int. 2012;32:476–86.
- Chen Y, Xu HX, Wang LJ, Liu XX, Mahato RI, Zhao YR. Metaanalysis: IL28B polymorphisms predict sustained viral response in HCV patients treated with pegylated interferon-alpha and ribavirin. Aliment Pharmacol Ther. 2012;36:91–103.
- Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, et al. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109–16.
- Stattermayer AF, Stauber R, Hofer H, Rutter K, Beinhardt S, Scherzer TM, et al. Impact of IL28B genotype on the early and sustained virologic response in treatment-naive patients with chronic hepatitis C. Clin Gastroenterol Hepatol. 2011;9:344–50 e2.
- Schreiber J, Moreno C, Garcia BG, Louvet A, Trepo E, Henrion J, et al. Meta-analysis: the impact of IL28B polymorphisms on rapid and sustained virological response in HCV-2 and -3 patients. Aliment Pharmacol Ther. 2012;36:353–62.
- Chinnaswamy S. Genetic Variants at the IFNL3 locus and their association with hepatitis C virus infections reveal novel insights into host-virus interactions. J Interferon Cytokine Res. 2014;34:479–97.
- Sarin SK, Kumar CK. Treatment of patients with genotype 3 chronic hepatitis C—current and future therapies. Liver Int. 2012;32 Suppl 1:141–5.
|
|
|
 |
|
|