|
|
|
|
 |
 |
| |
 |
|
|
Original Articles |
|
|
|
|
|
Keywords :
Shigella spp. serogroup distribution, Ile-Ife |
|
|
|
Abdulrasheed Abdu,1 Aaron O Aboderin,2 Jerome B Elusiyan,3 DO Kolawole,4 Adebayo Lamikanra5
Department of Medical
Microbiology and Parasitology,1
Faculty of Basic Medical Sciences,
College of Health Sciences,
Niger Delta University,
Wilberforce Island, Bayelsa-State,
Department of Medical
Microbiology and Parasitology,2
Faculty of Basic Medical Sciences,
College of Health Sciences,
Department of Paediatrics,3 Faculty
of Clinical Sciences, College of Health Sciences,
Department of Microbiology,4
Faculty of Sciences,
Department of Pharmaceutics,5
Faculty of Pharmacy,
Obafemi Awolowo University,
Ile-Ife, Osun-State, Nigeria
Corresponding Author:
Dr Abdu Abdulrasheed
Email: abdulsoul@gmail.com
DOI:
http://dx.doi.org/10.7869/tg.121
Abstract
Background: Shigellosis is endemic throughout the world and Shigella spp. is among the most common pathogens responsible for bacterial diarrhoeal diseases. Death attributed to shigellosis is common in developing countries, where affected populations are immunologically compromised due to poor nutrition and background infections.
Aim: To investigate the serogroup distribution of Shigella spp. recovered from clinically diagnosed cases of gastroenteritis and acute diarrhoea among children (0–5 years) in Ile-Ife, southwest Nigeria between September 2003 and September 2006.
Methods: The isolates were identified and characterized biochemically and serologically. Results: Out of 102 Shigella isolates identified, 45 (44%) were S. flexneri, 26 (25%) were S. dysenteriae, 19 (19%) were S. boydii, 6 (6%) were S. sonnei and 6 (6%) were untypable strains.
Conclusions: We conclude that Shigella serogroups can be considered an important aetiological agent of acute diarrhoea and mortality among children in Ile-Ife, southwest Nigeria.
|
48uep6bbphidvals|606 48uep6bbph|2000F98CTab_Articles|Fulltext Shigella is named after Kiyoshi Shiga, who was the first scientist to isolate Shigella dysenteriae type 1 in the year 1896 during a large epidemic of dysentery in Japan.[1] Shigellosis is a collective description for infectious diseases caused by members of the bacterial genus Shigella, whose transmission occurs mostly via the faecal–oral route. Most Shigella outbreaks are associated with over-crowding, poor personal hygiene and abysmal conditions in daycare centres, nursing homes, custodial institutions, cruise ships, aboriginal reservations and refugee camps[2] with contaminated food or water serving as the vehicle for infection. The infectious dose of Shigella spp. is low, with 10–100 bacteria/mL sufficient to cause the disease.[3] In developed countries, shigellosis transmission has also been linked to oro-genital and oro-anal sexual contact between men[4] and more recently with underlying human immunodeficiency virus (HIV) infection.[5]
Shigellosis is endemic throughout the world and it is among the most common pathogens associated with bacterial diarrhoeal diseases.6 It is particularly common in developing countries where affected populations are immunologically compromised due to poor nutrition and background infections, leading to high morbidity attributed to shigellosis.[1,4,7] Although epidemic Shigella dysentery is the most serious manifestation of Shigella infection in developing countries, the majority of Shigella infections are due to endemic shigellosis. Epidemiological reports have shown that shigellosis is responsible for approximately 165 million cases annually, of which 163 million (98.8%) are in developing countries and 1.5 million in the industrialized countries.[7] It has also been estimated that between 600,000 and 1.1 million (mean 850,000) people die annually from Shigella infection[7–8] and nearly 580,000 cases of shigellosis are reported among travellers from industrialized nations travelling to developing countries.[9] The genus Shigella comprises four subgroups that historically have been considered species, and these four Shigella spp. are recognized as pathogenic to humans. Subgroup A is referred to as S. dysenteriae, subgroup B as S. flexneri, subgroup C as S. boydii and subgroup D as S. sonnei.[10] Both S. sonnei and S. boydii are usually associated with mild illness of short duration in which the stool may be watery or bloody.[11] S. flexneri is generally more severe, lasts longer, causes dysentery more frequently and is the principal cause of endemic shigellosis in many developing countries.[12] Of all serotypes of Shigellae, particularly type 1 S. dysenteriae, attracts special attention for its endemic and epidemic-causing potential as well as its high attack rate, high case-fatality rate, and various complications.[13]
Significant changes in the global epidemiology of Shigellae spp. have been observed over the past decades of the 20th century. In industrialized regions, S. dysenteriae was first replaced by S. flexneri, and then S. sonnei;[14] and has been predominantly involved in common source sporadic outbreaks[12] accounting for a prevalence rate of 77%.[15] S. flexneri remains the leading cause of shigellosis in most of the developing world[16] and has been reported in approximately 10% of all diarrhoeal episodes among children younger than 5 years of age.[12] According to Peirano et al.[6] the frequency of S. flexneri, S. sonnei, S. boydii and S. dysenteriae was 60%, 15%, 6% and 6%, respectively (30% of S. dysenteriae cases were type 1) in developing countries; and 16%, 77%, 2% and 1%, respectively in the developed nations. Evidence has shown that in developing nations, the predominant serotype of S. flexneri is 2a, followed by 1b, 3a, 4a and 6.[17]
The current study was undertaken to investigate the serogroup distribution of Shigella spp. isolated from clinically diagnosed cases of gastroenteritis and acute diarrhoea in Ile- Ife—a semi-urban area in southwest Nigeria.
Methods
Patient enrolment and sample collection was carried out from September 2003 to September 2006. A total of 300 consecutive stool specimens were collected from children with acute diarrhoea admitted to the Children’s Emergency Ward at Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC), Ile-Ife, southwest Nigeria.
Bacteriological procedures
Rectal swabs or fresh stool samples were collected into Stuart transport medium (Oxoid, England) and processed within 2 hours of collection following the procedure described by Chompook et al.[18] and Iwalokun et al.[19] The samples were simultaneously inoculated directly on MacConkey agar (Oxoid, England) and Salmonella–Shigella agar, and were incubated at 37 °C overnight. Overnight cultures were tested for enteropathogens by employing standard techniques as outlined by Bopp et al.[20]
Biochemical characterization
Colonies morphologically resembling Shigellae were further subcultured and identified by biochemical reactions on API 20E.
Serogrouping
Serogroups were determined by using commercially available antisera kits (Sanofi Diagnostics Pasteur, France and Pro-Lab Diagnostics, England). Strains were subcultured on Salmonella– Shigella agar (Oxoid, England) and serotyping was performed after ~18 hours of incubation at 37 ÚC, by using the slide agglutination tests as described by the manufacturers.
Results
Of the 300 diarrhoeal stool samples screened, a total of 102 (34%) Shigella strains were identified. Clinical symptoms and other characteristics of the 102 patients with Shigella spp. isolated from their stool specimen are summarized in Table 1. Fever, blood in stool, and abdominal pain were significantly higher (p<0.002, p<0.0001 and p<0.05, respectively), and mucus in stool was significantly lower (p<0.04) in patients whose specimen yielded Shigella spp. as compared to the rest of the patients. Other characteristics and clinical symptoms did not show a significant difference between the above-mentioned groups. Among the patients with Shigella, fever, blood in stool, anorexia and abdominal pain were significantly higher among those who were aged e”37 months (p<0.001), while vomiting was more prevalent among those aged <37 months (p<0.001). The physicians prescribed intravenous fluid and antibiotics to all the patients.
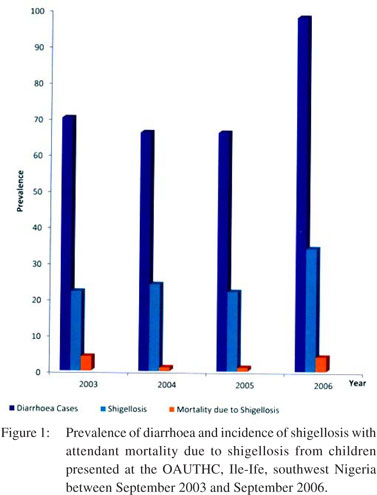
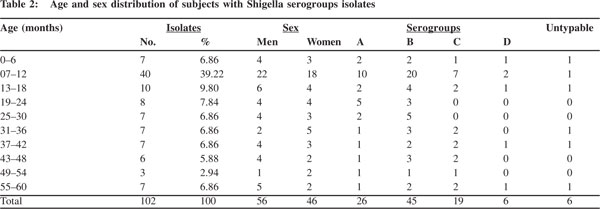 Figure 1 shows the prevalence of diarrhoea and the incidence of shigellosis with attendant mortality due to shigellosis, among the children enrolled in this study. Of the 102 children infected with Shigella spp., 10 (9.8%) were lost to their illness. Age-distributed mortality rate revealed that 2 (14.3%), 2 (11.1%) and 4 (8.5%) deaths occurred in children of 25–36 months, 13–24 months and 0–12 month(s) of age, respectively (table not shown). Fatality rate obtained was highest in patients with S. dysenteriae (70%) with a mortality rate of 30% associated with S. flexneri. None of the children infected with either S. boydii or S. sonnei died.
Figure 1 shows the prevalence of diarrhoea and the incidence of shigellosis with attendant mortality due to shigellosis, among the children enrolled in this study. Of the 102 children infected with Shigella spp., 10 (9.8%) were lost to their illness. Age-distributed mortality rate revealed that 2 (14.3%), 2 (11.1%) and 4 (8.5%) deaths occurred in children of 25–36 months, 13–24 months and 0–12 month(s) of age, respectively (table not shown). Fatality rate obtained was highest in patients with S. dysenteriae (70%) with a mortality rate of 30% associated with S. flexneri. None of the children infected with either S. boydii or S. sonnei died.
The frequency distribution of serogroups A, B, C and D is shown in Table 2. The serogroups prevalence of the 102 Shigella isolates was as follows: S. flexneri strains were the most frequently encountered [n=45 (44.12%)], while S. dysenteriae, S. boydii and S. sonnei accounted for 26 (25.49%), 19 (18.63%) and 6 (5.88%) isolates, respectively. Six (5.88%) of the isolates were untypeable.
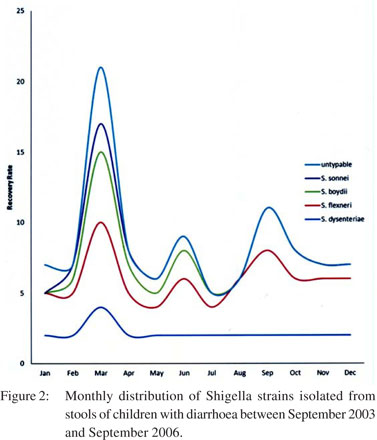
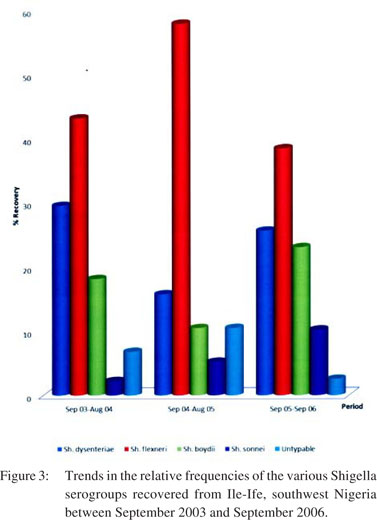
Figure 2 shows the month-wise distribution to highlight the number of Shigella serogroups isolated per season. The highest incidence of Shigella infections was found in March (20.59%), while the lowest numbers were observed in July (4.90%). Figure 3, on the other hand, illustrates the annual trend of Shigella serogroup isolation.
Discussion
In the present study, we investigated the serogroup distribution of 102 isolates of Shigella isolated in Ile-Ife, a semi-urban settlement, in southwest Nigeria, from . Shigellosis, like in many other developing countries, is among the main health problems in Ile-Ife and accounts for about 34% of the disease incidence in the study area.
The speciation showed that serogroup B—S. flexneri is the most common species [45 (44.1%)] among the total cases of shigellosis observed during the study period. Serogroup A— S. dysenteriae and serogroup C—S. boydii were identified in 26 (25.5%) and 19 (18.6%) cases, respectively; while 6 (5.9%) cases were caused by serogroup D—S. sonnei. Six (5.9%) of the isolates were serologically untypeable with the available commercial antisera. This pattern of shigellosis indicates that S. flexneri is the predominant and most active serogroup in Ile- Ife which is consistent with the patterns of isolates reported from other parts of Nigeria by Enabulele et al.,[5] Iwalokun et al.,[19] Egah et al.,[21] and Ogunlesi et al.[22] It is pertinent to point out that the order of serogroups distribution observed in this study had also been reported earlier in several studies done elsewhere, especially in other parts of Africa as reported by Shapiro et al. from Rwanda,[23] Kenya;[24] Yismaw et al. from Ethiopia[25] and Opintam and Newman from Ghana.[26] The observed results are also in keeping with the prevalence data reported from other developing countries,[18,27,28] thus emphasizing the continued predominance of S. flexneri in developing nations. Our study findings indicate that S. flexneri is the predominant Shigella spp. recovered from patients with acute diarrhoea in developing countries with an isolation rate of 50%–70% of all Shigella isolates recovered.[6,29] This result is, however, in contrast with studies from developed countries where S. sonnei is the predominant species isolated in Shigella-associated diarrhoea, and S. flexneri is the second most prevalent Shigella isolate.[30] For instance, S. sonnei has been documented to account for 64% of all Shigella spp. isolated in the USA,[31] and 80% in European countries.[32] This is in line with the reports that S. sonnei is dominant in more economically successful countries and the epidemiological decrease in the proportion of S. flexneri and S. dysenteriae strains in the industrialized countries has been attributed to the improvement in hygiene and living conditions in these countries.6 In addition, Naik[33] has attributed to improved sanitary conditions the progressive substitution of the most pathogenic Shigella spp. (e.g. S. flexneri) by less virulent ones (e.g. S. sonnei). These observations imply that the circulation of the more virulent Shigella spp. tends to depreciate with improvements in hygiene and living conditions as in developed countries, to be replaced by the less virulent Shigella strains and a significantly lower prevalence of Shigella-associated diarrhoea.[34]
Unlike other places across the world, where surveillance of Shigella species has revealed a distinct shift in the circulating species associated with socioeconomic development over time,[35] S. flexneri continues to remain the predominant species in Nigeria, indicating no significant improvement in socioeconomic living conditions of our populace over the past two decades.[19] Although we have not included a detailed description and comparison of the sanitary condition of our subjects’ living conditions, it would suffice to indicate that sanitary conditions are inevitably abysmal in such communities lacking a secure water supply and proper waste disposal system. It is also worth noting that the manually dug shallow wells, which are the major source of drinking water in these communities, are highly vulnerable to contamination with faecal pathogens, including Shigella spp. Acute shortage of clean potable water forces such communities to be parsimonious with their water supply, thus neglecting the basic tenets of hygiene including hand-washing, which can interrupt transmission of pathogens such as Shigella.[22] Most of the subjects in such communities rely on ponds, streams and other surface sources of water for bathing, washing clothes and maintaining livestock thus contaminating these water sources with infectious human and animal wastes. Shigella infection is contracted due to poor personal hygiene, consuming contaminated food and water and person-to-person contact. Similar to other enteric pathogens, Shigella can also be transferred by flies,[36] fingers and fomites.[6]
The results of this study are also in agreement with the results of studies from various parts of Nigeria, which have shown that S. dysenteriae is the second most prevalent species in Lagos and Ilesa, southwest Nigeria,[19,22] in Calabar, south Nigeria[5] and in Enugu, southeast Nigeria;[37] but these are in contrast to the study from north-central Nigeria, which detected S. boydii as the second most common species in Jos.[21] This pattern of geographical species diversity among Shigella isolates within a country has been reported in earlier studies carried out in India[38] and Brazil.[6,39]
This study has shown that S. boydii serogroup C is also an important aetiological agent in Ile-Ife, southwest Nigeria and its isolation rate [19 (18.6%)] is similar to [11 (17.7%)] earlier reports by Iwalokun et al.[19] from Lagos. These observations also support the report that S. boydii accounts for several cases of human disease in developing countries.[40] Recent reports from Canada indicate that there has been a steady increase in S. boydii isolation rate starting in 1999 and this species is being most frequently recovered from individuals travelling to developing countries such as Cuba, Ethiopia, India, Guatemala and Mexico.[7]
The present study showed a very low occurrence of S. sonnei infections [6 (5.9%)]. This is in line with the report that S. sonnei occurs more commonly in developed countries, often emerging at epidemic levels or as sporadic outbreaks.[21] S. sonnei is responsible for about 77% of Shigella infections in industrialized countries.[27]
Conclusions
Based on our findings in this study, we conclude that Shigella serogroups can be considered an important aetiological agent of acute diarrhoea and mortality among children in Ile-Ife, southwest Nigeria. Given the gravity of the situation, it is imperative to prevent this infection through promotion of health education among food-handlers as well as close attention should be given to the improvement of good hygiene practices among the general populace.
References
- Mammina C, Ranjbar R. Shigellosis: disease and epidemiology (2008). Online open learning on enteric pathologies course. Available at http://www.aclisassari.com/acli-openlearning/ uploads/Shigella.ppt.
- Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): 2002 Human Isolates Final Report. Atlanta, Georgia: US Department of Health and Human Services, CDC. 2004. Available at http://www.cdc.gov/narms/annual/2002/2002ANNUALREPORTFINAL.pdf.
- Travejo RT, Abbott SL, Wolfe MI, Meshulam J, Yong D, Flore GR. An untypable Shigella flexneri strain associated with an outbreak in California. J Clin Microbiol. 1999;37:2352–3.
- O’Sullivan B, Delpech V, Pontivivo G, Karagiannis T, Marriott D, Harkness J, et al. Shigellosis linked to sex venues, Australia. Emerg Infect Dis. 2002;8:862–4.
- Enabulele IO, Yah SC, Yusuf EO, Eghafona NO. Emerging quinolones resistant transfer genes among gram-negative bacteria, isolated from faeces of HIV/AIDS patients attending some clinics and hospitals in the city of Benin, Edo State, Nigeria. Online J Health Allied Scs. 2006;5:1–9.
- Peirano G, Souza FS, Rodrigues DP, Shigella Study Group. Frequency of serovers and antimicrobial resistance in Shigella spp. from Brazil. Mem Inst Oswaldo Cruz. 2006;101:245–50.
- Woodward DL, Clark CG, Caldeira RA, Ahmed R, Soule G, Bryden L, et al. Identification and characterization of Shigella boydii 20 serovar nov., a new and emerging Shigella serotype. J Med Microbiol. 2005;54:741–8.
- World Health Organization. Diarrhoeal disease due to Shigella disease. In: Vaccines, immunization and biologicals. Geneva: WHO, 1998;1–5.
- Health Protection Agency. Imported Infections, England and Wales: July to September, 2006. Health Protect Rep. 2007;1:1–15.
- Talukder KA, Mondol AS, Islam AM, Islam Z, Dutta DK, Khajanchi BK, et al. A novel serovar of Shigella dysenteriae from patients with diarrhoea in Bangladesh. J Med Microbiol. 2007;56:654–8.
- Keusch GT, Bennish LM. Shigellosis. In: Evans AS, Brachman PS (eds). Bacteria infections of humans: epidemiology and control. 3rd ed. New York, USA: Plenum; 1998:631–56.
- Niyogi SK. Shigellosis. J Microbiol. 2005;43:133–43.
- World Health Organization. The management of bloody diarrhoea in young children. Geneva: WHO 1994 (unpublished document WHO/CDD/94.49).
- Ashkenazi S, Amir J, Waisman Y, Rachmel A, Garty B, Samara Z, et al. A randomized double-blind study comparing cefixime and trimethoprim-sulfamethoxazole in the treatment of childhood shigellosis. J Paediatr. 1993;123:817–21.
- Talukder KA, Islam Z, Dutta DK, Islam AM, Khajanchi BK, Azmi IJ, et al. Antibiotic resistance and genetic diversity of Shigella sonnei isolated from patients with diarrhoea between 1999 and 2003 in Bangladesh. J Med Microbiol. 2006;55:1257–63.
- Subekti D, Oyofo A, Tjaniadi P, Corwin AL, Larasati W, Putri M, et al. Shigella spp. surveillance in Indonesia: the emergence or reemergence of S. dysenteriae. Emerg Infect Dis. 2001;7:137–40.
- Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–66.
- Chompook P, Samosornsuk S, von Seidlein L, Jitsanguansuk S, Sirima N, Sudjai S, et al. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83:739–46.
- Iwalokun BA, Gbenle GO, Smith SI, Ogunledun A, Akinsinde KA, Omonigbehin EA. Epidemiology of shigellosis in Lagos, Nigeria: trends in antimicrobial resistance. J Health Popul Nutr. 2001;19:183–90.
- Bopp CA, Brenner FW, Fields PL, Wells JG, Strockbine NA. Escherichia, Shigella, and Salmonella. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (eds). Manual of Clinical Microbiology. 8th ed. Washington DC: American Society of Microbiology; 2003:654–71.
- Egah DZ, Banwat EB, Audu ES, Allanana JA, Danung ML, Damen JG, et al. Multiple drug resistant strain of Shigella isolated in Jos, central Nigeria. Niger Postgrad J Med. 2003;10:154–6.
- Ogunlesi TA, Okeniyi JAO, Oyedeji OA, Oseni SBA, Oyelami OA, NjoKanma OF, et al. Childhood dysentery in Ilesa, Nigeria: the unusual role of Entamoeba histolytica. Internet J Trop Med. 2005;2:1–8.
- Shapiro RL, Kumar L, Phillips-Howard P, Wells JG, Adcock P, Brooks J, et al. Antimicrobial-resistant bacterial diarrhoea in rural western Kenya. J Infect Dis. 2001;183:1701–4.
- Brooks JT, Shapiro RL, Kumar L, Wells JG, Phillips-Howard PA, Shi YP, et al. Epidemiology of sporadic bloody diarrhea in rural western Kenya. Am J Trop Med Hyg. 2003;68:671–7.
- Yismaw W, Negeri C, Kassu A. A five-year antimicrobial resistance pattern observed in Shigella species isolated from stool samples in Gondar University Hospital, northwest Ethiopia. Ethiop J Health Dev. 2006;20:194–8.
- Opintan JA, Newman MJ. Distribution of serogroups and serotypes of multiple drug resistant Shigella isolates. Ghana Med J. 2007;41:8–29.
- Talukder KA, Islam AM, Khajanchi BK, Dutta DK, Islam Z, Safa A, et al. Temporal shifts in the dominance of serotypes of Shigella dysenteriae from 1999 to 2002 in Dhaka, Bangladesh. J Clin Microbiol. 2003;41:5053–8.
- Pazhani GP, Ramamurthy T, Mitra U, Bhattacharya SK, Niyogi SK. Species diversity and antimicrobial resistance of Shigella spp. isolated between 2001 and 2004 from hospitalized children with diarrhoea in Kolkata (Calcutta), India. Epidemiol Infect. 2005;133:1089–95.
- Fullá N, Prado V, Duran C, Lagos R, Levine MM. Surveillance for antimicrobial resistance profiles among Shigella species isolated from a semirural community in the northern administrative area of santiago, Chile. Am J Trop Med Hyg. 2005;72:851–4.
- Essers B, Burnens AP, Lanfranchini FM, Somaruga SG, von Vigier RO, Schaad UB. Acute community-acquired diarrhea requiring hospital admission in Swiss children. Clin Infect Dis. 2000;31:192–6.
- Sivapalasingam S, Nelson JM, Joyce K, Hoekstra M, Angulo FJ, Mintz ED. High prevalence of antimicrobial resistance among Shigella isolates in the United States tested by the National Antimicrobial Resistance Monitoring System from 1999 to 2002. Antimicrob Agents Chemother. 2006;50:49–54.
- Ozmert EN, Gokturk B, Yurdakok K, Yalcin SS, Gur D. Shigella antibiotic resistance in central Turkey: comparison of the years 1987–1994 and 1995–2002. J Paediatr Gastroenterol Nutr. 2005;40:359–62.
- Naik DG. Prevalence and antimicrobial susceptibility patterns of Shigella species in Asmara, Eritrea, northeast Africa. J Microbiol Immunol Infect. 2006;39:392–5.
- Koubek T, Herben T. Effect of systematic diseases in clonal integration: modelling approach. Evol Ecol. 2008;22:449–60.
- Dutta S, Rajendran K, Roy S, Chatterjee A, Dutta P, Nair GB, et al. Shifting serotypes, plasmid profile analysis and antimicrobial resistance pattern of Shigellae strains isolated from Kolkata, India during 1995-2000. Epidemiol Infect. 2002;129:235–43.
- Ugbogu OC, Nwachukwu NC, Ogbuagu UN. Isolation of Salmonella and Shigella species from house flies (Musca domestica) in Uturu, Nigeria. Afr J Biotechnol. 2006;5:1090–91.
- Ozumba UC. Antimicrobial susceptibility pattern and serogroup distribution of Shigella species at Enugu. Nig Postgrad Med J. 1997;4:1–3.
- Taneja N, Mohan B, Khurana S, Sharma M. Antimicrobial resistance in selected bacterial enteropathogens in north India. Indian J Med Res. 2004;120:39–43.
- Diniz-Santos DR, Santana JS, Barretto JR, Andrade MG, Silva LR. Epidemiological and microbiological aspects of acute bacterial diarrhea in children from Salvador, Bahia, Brazil. Braz J Infect Dis. 2005;9:77–83.
- Janda JM, Abbott SL. The genus Shigella. In: The Enterobacteria. Philadelphia, PA, USA: Lippincott-Raven; 1998:66–79.
|
|
|
 |
|
|